Scientists Map 37,000 Years of Human Infectious Disease in Landmark Effort to “Help Us Prepare For the Future” – The Debrief

Report on Ancient Pathogen Genomics and Implications for Sustainable Development Goals
Introduction: Aligning Historical Disease Research with Global Health Targets
A landmark study published in Nature provides critical insights into the historical spread of infectious diseases, directly informing strategies related to the United Nations Sustainable Development Goals (SDGs), particularly SDG 3 (Good Health and Well-being). Research conducted as a partnership between the University of Cambridge and the University of Copenhagen (a testament to SDG 17, Partnerships for the Goals) analyzed ancient human DNA to reveal how animal domestication and human migration patterns shaped the landscape of human pathogens. The findings underscore the deep historical roots of public health challenges and offer valuable data for future disease prevention and control.
Study Methodology and Scope
The research represents the most extensive analysis of ancient pathogen DNA ever completed. The study’s framework was built upon:
- Data Set: DNA extracted from the bones and teeth of over 1,300 ancient humans.
- Geographic Scope: Samples collected across Eurasia.
- Temporal Range: The oldest samples date back 37,000 years.
- Research Focus: To identify the spatiotemporal distribution of 214 known human pathogens and understand their origins and spread.
Key Findings: The Intersection of Human Activity and Disease Emergence
The Neolithic Transition and its Impact on SDG 3 and SDG 15
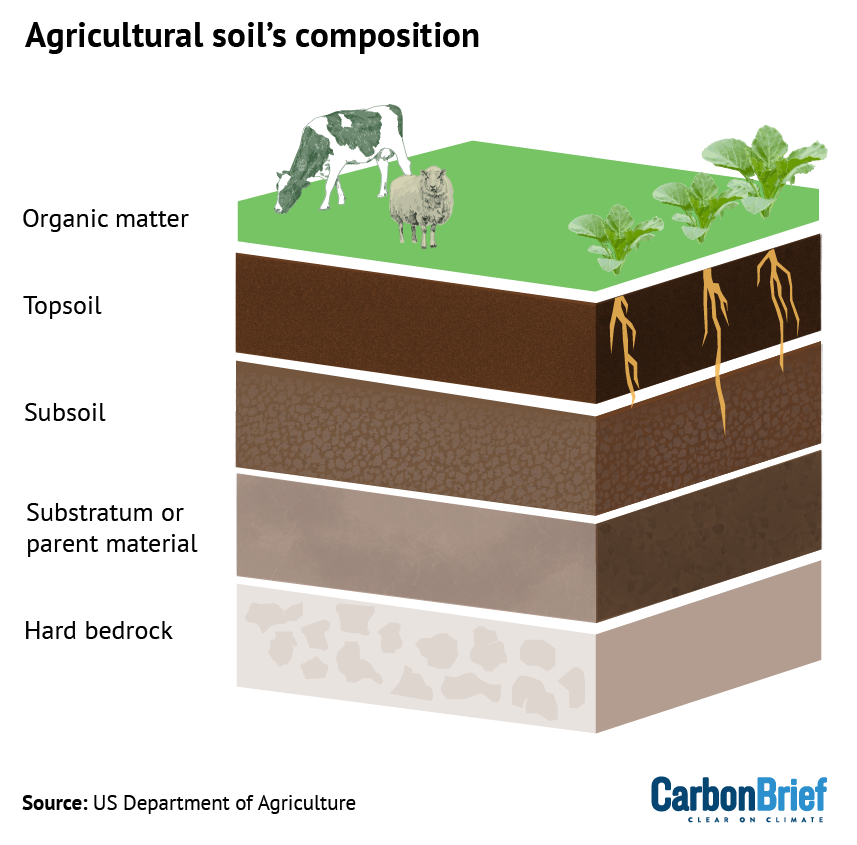
A central finding is that the transition to agriculture and the domestication of animals, beginning approximately 6,500 years ago, marked a new era for infectious diseases. This close cohabitation created pathways for zoonotic transmission, where pathogens jump from animals to humans. This directly relates to SDG 15 (Life on Land), which emphasizes the importance of managing ecosystems and halting biodiversity loss, as the human-animal interface remains a primary source of emerging infectious diseases. The study confirms that this interface was a critical factor in the rise of pathogens that continue to threaten SDG 3 (Good Health and Well-being).
Timeline of Ancient Pathogen Detection
The study successfully traced several significant pathogens to their earliest known appearances in the human genome. These findings are crucial for understanding the long-term evolution of diseases targeted by public health initiatives under SDG 3.
- Corynebacterium diphtheriae (Diphtheria): Evidence found dating back 11,100 years.
- Hepatitis B Virus: Traced back 9,800 years.
- Zoonotic Pathogens (e.g., Yersinia pestis): Began emerging approximately 6,500 years ago, with prevalence increasing 1,500 years later.
- Plasmodium vivax (Malaria): Identified in samples from around 4,200 years ago.
- Leprosy: Emerged approximately 1,400 years ago.
Human Migration and its Role in Pathogen Dispersal
Migration as a Disease Vector and Relevance to SDG 11
The research highlights that large-scale human migration was a key mechanism for introducing and spreading pathogens across vast regions. A significant rise in zoonotic pathogens around 5,000 years ago directly coincides with the migration of Yamnaya herders from the Pontic Steppe into northwestern Europe. This historical dynamic offers a parallel to modern challenges in global health, connecting to SDG 11 (Sustainable Cities and Communities). Understanding how population movements impact disease transmission is vital for building resilient and healthy communities, particularly in an era of unprecedented global mobility.
Modern Applications for Achieving Global Health Security
Informing Future Vaccine Development and Disease Preparedness
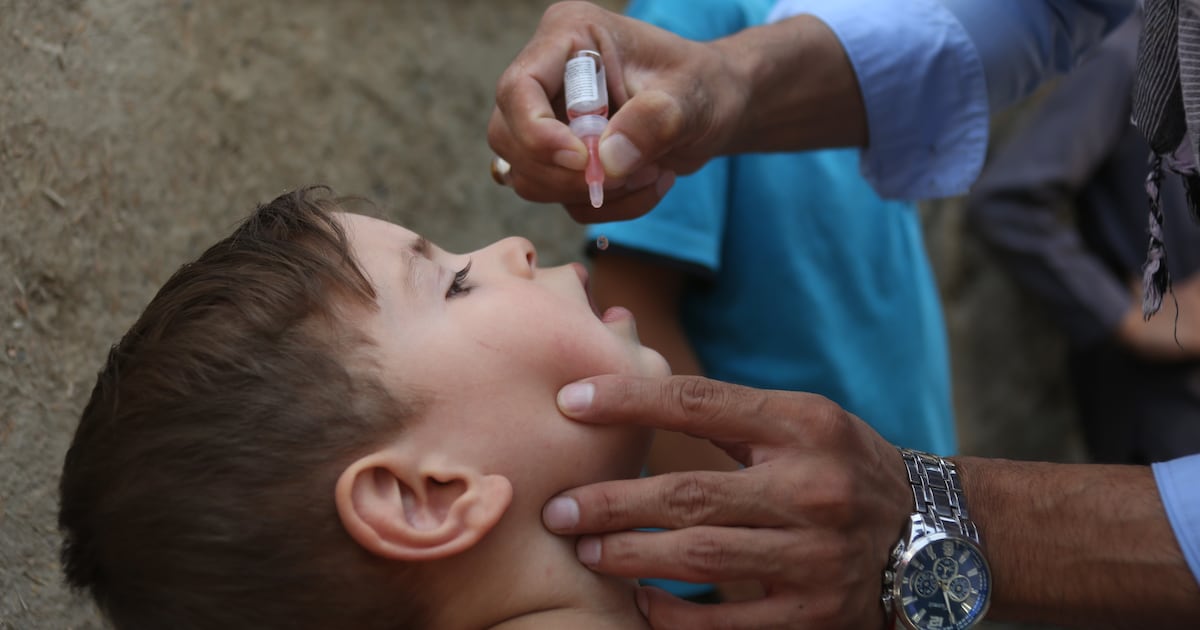
The study’s implications extend far beyond historical understanding. By mapping the long-term evolution of pathogens, researchers can better anticipate future mutations and emergences. This historical knowledge is a critical tool for achieving SDG 3, Target 3.b, which calls for supporting the research and development of vaccines and medicines.
- Predicting Mutations: Understanding how pathogens like those responsible for plague or diphtheria have evolved can help scientists predict which mutations are likely to reemerge.
- Vaccine Efficacy: This research allows for testing whether current vaccines provide adequate coverage against ancient and potentially reemergent strains, guiding the development of new vaccines.
- Preparing for Zoonosis: As noted by the study’s authors, many emerging infectious diseases are expected to be zoonotic. This historical data reinforces the need for a “One Health” approach, integrating human, animal, and environmental health to prevent future pandemics, a core tenet of modern public health strategy aligned with the SDGs.
This collaborative research effort, detailed in the Nature article “The spatiotemporal distribution of human pathogens in ancient Eurasia,” provides an essential historical foundation for tackling the modern challenges outlined in the Sustainable Development Goals, reinforcing the link between our past and our collective future health.
Which SDGs are addressed or connected to the issues highlighted in the article?
SDG 3: Good Health and Well-being
- The article’s core subject is the history and spread of infectious diseases, including plague, diphtheria, Hepatitis B, malaria, and leprosy. It directly connects this historical understanding to modern public health challenges, stating that the research could help “bolster future efforts to develop vaccines” and aid in understanding how diseases emerge. This aligns with the goal of ensuring healthy lives and promoting well-being for all at all ages.
SDG 11: Sustainable Cities and Communities
- The article highlights how large-scale human migration was a key factor in the spread of pathogens. It also notes that disease outbreaks “may have contributed to population collapse,” which directly relates to the resilience and sustainability of human settlements and communities against disasters, including biological ones like epidemics.
SDG 15: Life on Land
- The research points to the “transition to farming and animal husbandry” as the period when zoonotic diseases began to emerge through transmission from animals to humans. This connection between human activities, interaction with terrestrial ecosystems and animals, and the rise of disease is relevant to managing ecosystems sustainably to prevent future zoonotic spillovers.
SDG 17: Partnerships for the Goals
- The article explicitly states that the study was “a collaboration between researchers at the University of Cambridge and the University of Copenhagen.” This international scientific partnership to address a global health challenge is a direct example of the collaboration required to achieve the SDGs.
What specific targets under those SDGs can be identified based on the article’s content?
SDG 3: Good Health and Well-being
-
Target 3.3: By 2030, end the epidemics of AIDS, tuberculosis, malaria and neglected tropical diseases and combat hepatitis, water-borne diseases and other communicable diseases.
- The article provides historical context for several of these diseases, including “Hepatitis B virus,” “malaria (Plasmodium vivax),” and “Leprosy.” The research aims to understand their origins and mutations to better combat them today.
-
Target 3.b: Support the research and development of vaccines and medicines for the communicable and non-communicable diseases.
- The article directly supports this target by stating that the research is “important for future vaccines, as it allows us to test whether current vaccines provide sufficient coverage or whether new ones need to be developed due to mutations.”
-
Target 3.d: Strengthen the capacity of all countries… for early warning, risk reduction and management of national and global health risks.
- The research aims to help us “prepare for the future” by understanding how ancient pathogens emerged and spread. This knowledge directly contributes to the capacity for risk reduction and management of future disease outbreaks, especially since “Many of the newly emerging infectious diseases are predicted to originate from animals.”
SDG 11: Sustainable Cities and Communities
-
Target 11.5: By 2030, significantly reduce the number of deaths and the number of people affected… caused by disasters.
- The article discusses how ancient infections may have “contributed to population collapse.” Understanding the dynamics of disease spread, which can be considered a biological disaster, is crucial for mitigating their impact and reducing deaths in modern populations.
SDG 17: Partnerships for the Goals
-
Target 17.6: Enhance… international cooperation on and access to science, technology and innovation.
- The study itself, described as “a collaboration between researchers at the University of Cambridge and the University of Copenhagen,” is a direct manifestation of this target. The publication of their findings in the journal Nature further enhances access to this scientific knowledge.
Are there any indicators mentioned or implied in the article that can be used to measure progress towards the identified targets?
Target 3.3 (End epidemics)
- Implied Indicator: Incidence of communicable diseases. The article’s focus on tracing the origins of specific pathogens like Yersinia pestis (plague), Hepatitis B, and malaria implies that tracking and reducing the incidence of these diseases is the ultimate goal of such research.
Target 3.b (Support R&D for vaccines)
- Implied Indicator: Number of new vaccines developed or improved. The article’s conclusion that the knowledge “is important for future vaccines” and can determine if “new ones need to be developed” points directly to the development of new or more effective vaccines as a measurable outcome.
Target 3.d (Strengthen early warning and risk management)
- Implied Indicator: Strengthened capacity for disease surveillance and prediction. The statement that understanding past occurrences “can help us prepare for the future” implies that a key metric of progress is an improved ability to predict, surveil, and manage emerging infectious diseases.
Target 17.6 (International cooperation in science)
- Mentioned Indicator: Joint scientific publications and research collaborations. The article explicitly mentions the collaborative study between two international universities and its publication in Nature, which serves as a direct indicator of international scientific cooperation.
Table of SDGs, Targets, and Indicators
| SDGs | Targets | Indicators |
|---|---|---|
| SDG 3: Good Health and Well-being |
3.3: End the epidemics of… malaria and neglected tropical diseases and combat hepatitis… and other communicable diseases.
3.b: Support the research and development of vaccines and medicines. 3.d: Strengthen the capacity… for early warning, risk reduction and management of national and global health risks. |
Implied: Incidence of communicable diseases (malaria, hepatitis, leprosy).
Implied: Development and improvement of new vaccines. Implied: Strengthened capacity for disease surveillance and prediction. |
| SDG 11: Sustainable Cities and Communities | 11.5: Significantly reduce the number of deaths and the number of people affected… caused by disasters. | Implied: Reduced mortality and population impact from epidemics (biological disasters). |
| SDG 17: Partnerships for the Goals | 17.6: Enhance… international cooperation on and access to science, technology and innovation. | Mentioned: Joint scientific research collaborations and publications between international institutions (University of Cambridge and University of Copenhagen). |
Source: thedebrief.org

What is Your Reaction?
 Like
0
Like
0
 Dislike
0
Dislike
0
 Love
0
Love
0
 Funny
0
Funny
0
 Angry
0
Angry
0
 Sad
0
Sad
0
 Wow
0
Wow
0



























;Resize=805#)