Plasma proteomics for biomarker discovery in childhood tuberculosis – Nature
Report on Plasma Proteomics for Biomarker Discovery in Childhood Tuberculosis
Executive Summary
This report details a study aimed at addressing critical gaps in pediatric tuberculosis (TB) diagnostics, a major impediment to achieving Sustainable Development Goal 3 (SDG 3), which targets ending the TB epidemic by 2030. Delays in diagnosing TB in children contribute significantly to mortality, with current sputum-based tests performing poorly in this demographic. This research utilized high-throughput plasma proteomics and machine learning on a diverse cohort of 511 children from four low- and middle-income countries (LMICs). The study successfully identified four minimal protein biosignatures (comprising 3 to 6 proteins) that can distinguish TB from other respiratory diseases with high accuracy (AUCs of 0.87–0.88). These biosignatures meet the World Health Organization’s (WHO) minimum accuracy requirements for a TB screening test. The development of a non-sputum-based test directly supports SDG 3.3 (end epidemics of communicable diseases), SDG 3.2 (end preventable deaths of children under 5), and SDG 10 (reduce inequality) by providing a tool to improve health outcomes in a vulnerable and disproportionately affected population. The international collaboration also exemplifies SDG 17 (Partnerships for the Goals).
Introduction: Addressing SDG 3 through Advanced Pediatric TB Diagnostics
Tuberculosis remains a leading cause of death from an infectious disease globally, undermining progress towards SDG 3: Good Health and Well-being. Children bear a disproportionate burden, accounting for 12% of TB cases but 15% of deaths, primarily because 96% of fatal cases occur in children who have not initiated treatment. This diagnostic and treatment gap is a direct challenge to achieving SDG Target 3.3 (to end the TB epidemic) and SDG Target 3.2 (to end preventable deaths of children under five).
The primary obstacles in pediatric TB diagnosis are:
- Inability of children to reliably produce sputum for testing.
- Low sensitivity of microbiological tests due to the paucibacillary nature of pediatric TB.
Consequently, an estimated half of all children with TB are not reported or diagnosed, creating a significant public health crisis. The development of a non-sputum-based biomarker test is a global priority to close this gap, improve case detection, and ensure timely treatment, thereby advancing universal health coverage (SDG Target 3.8).
Methodology: A Global Partnership for Innovation (SDG 17)
This study represents a significant international collaboration, aligning with SDG 17: Partnerships for the Goals, by bringing together researchers and clinical cohorts from The Gambia, Peru, South Africa, and Uganda. This multi-continental approach ensures the developed biomarkers are robust and not specific to a single regional environment or comorbidity profile.
Cohort Characteristics and Ethical Framework
The study analyzed plasma samples from 511 children under evaluation for pulmonary TB. The cohort was designed to reflect real-world diagnostic challenges and address health inequalities (SDG 10) by including diverse and vulnerable populations.
- Patient Groups: Children were classified using NIH consensus definitions into Confirmed TB (n=133), Unconfirmed TB (n=120), and Unlikely TB (n=231, serving as controls with other respiratory infections). This design ensures biomarkers are specific to TB and not general inflammation.
- Demographics: The cohort included a median age of 4 years, 11.2% of children living with HIV, and 52.6% who were underweight, reflecting key risk groups often excluded from research.
High-Throughput Proteomics and Data Analysis
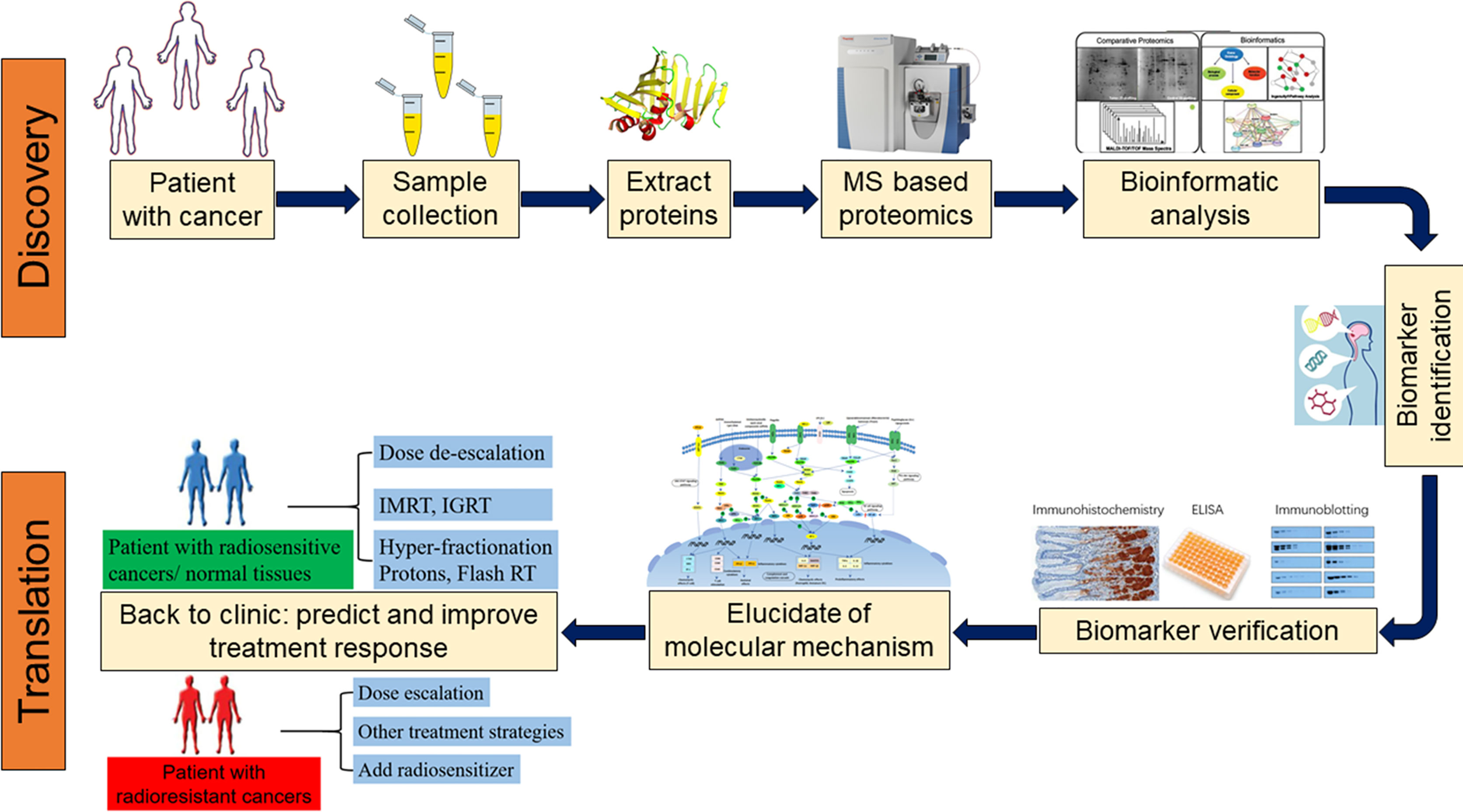
A high-throughput, data-independent acquisition mass spectrometry (DIA-PASEF) workflow was employed to analyze the plasma proteome from just 1µL of plasma. This efficient method quantified 859 proteins across the cohort. A machine learning approach, specifically a LASSO model followed by exhaustive combinatorial analysis, was used to identify the most parsimonious and effective protein biosignatures for diagnosing TB.
Key Findings: Identification of a Novel Biosignature for TB
The analysis yielded several crucial findings that pave the way for a new generation of pediatric TB diagnostics, directly contributing to the goals of SDG 3.
Differentiating TB from Other Respiratory Illnesses
A comparison between children with Confirmed TB and those with Unlikely TB (other respiratory diseases) identified 47 proteins with significantly different abundance levels. Key findings include:
- Upregulated Proteins: 17 proteins were upregulated in TB cases, including WARS1, a tryptophanyl t-RNA synthetase previously linked to adult TB, suggesting its importance in the host response.
- Downregulated Proteins: 30 proteins were downregulated.
- Pathway Analysis: Pathway enrichment analysis revealed significant dysregulation in pathways related to complement activation, a known feature of the immune response in TB.
Development of a Parsimonious Diagnostic Model
Using machine learning, the study identified four minimal biosignatures that performed with high accuracy. These models are a critical step towards developing a practical point-of-care test.
- 3-Protein Model: Met the WHO Target Product Profile (TPP) for a screening test (≥90% sensitivity at ≥70% specificity).
- 4-Protein Model: Included WARS1, APOM, TNC, and CD44.
- 5-Protein Model: Achieved 93% sensitivity at 70% specificity.
- 6-Protein Model: Achieved 96.7% sensitivity at 70% specificity.
The success of these minimal models (3-6 proteins) is vital for translation into a simple, cost-effective diagnostic tool suitable for use in resource-limited settings, which is essential for achieving equitable health outcomes under SDG 3 and SDG 10.
Performance and Specificity of the Biosignature
The derived biosignatures demonstrated high potential for clinical utility. When applied to the 115 “Unconfirmed TB” cases (children with clinical symptoms who were negative on standard tests but improved with treatment), the models supported a TB diagnosis in approximately 79% of cases. Furthermore, the biosignatures did not differentiate between healthy children and those with latent TB, indicating they are specific for active TB disease. This specificity is crucial for avoiding unnecessary treatment and ensuring resources are directed appropriately.
Discussion: Implications for Global Health and Sustainable Development
Contribution to Ending the TB Epidemic (SDG 3.3)
The identification of a robust, non-sputum-based protein biosignature is a landmark achievement. If developed into a point-of-care test, it could revolutionize pediatric TB management by:
- Reducing Diagnostic Delays: Enabling rapid diagnosis and treatment initiation.
- Closing the Case Detection Gap: Identifying the large number of children currently missed by conventional diagnostics.
- Lowering Child Mortality: Directly addressing the high death rate from untreated TB, contributing to SDG Target 3.2.
Reducing Health Inequalities (SDG 10)
This research actively works to reduce health inequalities by focusing on a vulnerable and neglected population: children with TB in LMICs. The inclusion of children with HIV and malnutrition ensures that the resulting diagnostic tool will be applicable to those at highest risk. By creating a tool that is effective in diverse settings, this work promotes equitable access to life-saving diagnostics.
Study Limitations and Future Directions
While promising, the biosignatures require further validation in prospective clinical trials to confirm their performance. The next critical step is the development of technologies, such as multiplexed point-of-care platforms, to translate these proteomic findings into a deployable test that can be used in clinics worldwide to achieve the ambitious targets set by the SDGs.
Conclusion: A Step Forward for Child Health and the SDGs
This study successfully leveraged untargeted proteomics and machine learning across a large, multi-national pediatric cohort to identify a minimal set of host protein biomarkers for childhood TB. The resulting 3- to 6-protein biosignatures meet WHO accuracy standards for a screening test and are specific for active disease. This work provides profound insights into the host immune response to pediatric TB and delivers a tangible, non-sputum-based solution that could dramatically reduce diagnostic delays and improve the management of TB in children globally. Ultimately, this research represents a significant scientific advancement in the fight against a major global health threat and a crucial step towards achieving the health-related Sustainable Development Goals.
Analysis of Sustainable Development Goals in the Article
1. Which SDGs are addressed or connected to the issues highlighted in the article?
-
SDG 3: Good Health and Well-being
- The article’s primary focus is on improving the diagnosis of Tuberculosis (TB), a major global infectious disease. It directly addresses the high mortality rate from TB, especially in children, stating it is the “leading cause of mortality from an infectious disease worldwide.” The research aims to develop a new diagnostic tool to reduce diagnostic delays, initiate treatment faster, and ultimately decrease deaths, which is central to ensuring healthy lives and promoting well-being. The article also notes the comorbidity with HIV, another major health challenge targeted by SDG 3.
-
SDG 17: Partnerships for the Goals
- The study is a multi-stakeholder and international collaboration. The research was conducted “across 4 countries” (The Gambia, Peru, South Africa, and Uganda) on two continents. This global partnership, involving various research institutions and clinical sites in Low- and Middle-Income Countries (LMICs), is essential for gathering diverse data and developing a robust solution applicable worldwide, embodying the spirit of strengthening the means of implementation and revitalizing global partnerships for sustainable development.
2. What specific targets under those SDGs can be identified based on the article’s content?
-
Target 3.2: End preventable deaths of newborns and children under 5 years of age.
- The article explicitly states that TB is a “leading cause of death in children” and that children suffer a “disproportionate burden.” It highlights that “96% of deaths are in children for whom treatment had not been initiated,” a problem directly caused by the failure to rapidly diagnose the disease. The development of a “non-sputum biosignature that could reduce delays in TB diagnosis” is a direct effort to prevent these deaths and reduce child mortality.
-
Target 3.3: By 2030, end the epidemics of AIDS, tuberculosis, malaria and neglected tropical diseases…
- The entire study is dedicated to combating the TB epidemic. It aims to close the “large case detection gap” in pediatric TB, which is a critical step toward ending the epidemic. The article also addresses the intersection of TB and HIV, noting that children with confirmed TB were “significantly more likely to be living with HIV,” aligning with the goal of tackling both epidemics.
-
Target 3.b: Support the research and development of vaccines and medicines for the communicable and non-communicable diseases that primarily affect developing countries…
- This research is a prime example of R&D for a new diagnostic tool for a communicable disease that disproportionately affects children in developing countries. The article states that the “development of non-sputum biomarker TB tests is a global priority.” The study uses advanced technologies like “high-throughput proteomics” and “machine learning” to create a novel biosignature, directly contributing to the scientific innovation needed to address these diseases.
3. Are there any indicators mentioned or implied in the article that can be used to measure progress towards the identified targets?
-
TB Mortality and Incidence Rates (related to Target 3.3)
- The article provides statistics that serve as baseline indicators, such as “10.8 million cases and 1.3 million deaths each year” from TB. It also specifies the pediatric burden, noting “12% of TB disease occurs in children, but children account for 15% of TB deaths.” A reduction in these numbers would indicate progress.
-
Case Detection Gap (related to Target 3.3)
- The article implies an indicator by stating there is a “large case detection gap where an estimated half of the children with TB disease… are not reported.” The success of the new diagnostic tool could be measured by its ability to close this gap, increasing the number of reported and treated cases.
-
Diagnostic Accuracy (related to Target 3.b)
- The article explicitly uses the World Health Organization’s (WHO) target product profile (TPP) as a key performance indicator. It states the goal is to achieve “≥ 90% sensitivity and ≥ 70% specificity.” The study’s success is measured against this indicator, reporting that the new biosignatures “all reach the minimum WHO target product profile accuracy thresholds” and achieve “AUCs of 0.87–0.88.”
4. Table of SDGs, Targets, and Indicators
| SDGs | Targets | Indicators |
|---|---|---|
| SDG 3: Good Health and Well-being |
3.2: End preventable deaths of newborns and children under 5 years of age.
3.3: End the epidemics of AIDS, tuberculosis, malaria… 3.b: Support the research and development of vaccines and medicines for communicable diseases that primarily affect developing countries. |
|
| SDG 17: Partnerships for the Goals | 17.6: Enhance North-South, South-South and triangular regional and international cooperation on and access to science, technology and innovation. |
|
Source: nature.com

What is Your Reaction?
 Like
0
Like
0
 Dislike
0
Dislike
0
 Love
0
Love
0
 Funny
0
Funny
0
 Angry
0
Angry
0
 Sad
0
Sad
0
 Wow
0
Wow
0























































.jpg?#)